Images photometry#
After an observation is done, a common need is to reduce and extract fluxes from raw FITS images.
In this tutorial you will learn how to process a complete night of raw data from any telescope by building a data reduction Sequence with prose.
Simualting data#
You can follow this tutorial on your own data or generate a synthetic dataset. As an example, let’s generate a light curve
import numpy as np
import matplotlib.pyplot as plt
from prose.simulations import simulate_observation
time = np.linspace(0, 0.15, 100) + 2450000
target_dflux = 1 + np.sin(time * 100) * 1e-2
plt.plot(time, target_dflux)
_ = plt.ylim(0.98, 1.02)
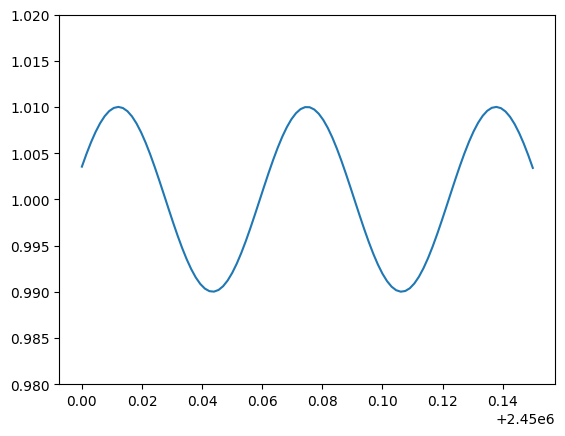
This might be the differential flux of a variable star. Let’s now simulate the fits images associated with the observation of this target:
# so we have the same data
np.random.seed(40)
fits_folder = "./tutorial_dataset"
simulate_observation(time, target_dflux, fits_folder)
100%|██████████| 100/100 [00:03<00:00, 27.58it/s]
here prose simulated comparison stars, their fluxes over time and some systematics noises.
Explore FITS#
The first thing we want to do is to see what is contained within our folder. For that we instantiate a FitsManager object on our folder to describe its content
from prose import FitsManager
fm = FitsManager(fits_folder, depth=2)
fm
| date | telescope | filter | type | target | width | height | files | |
|---|---|---|---|---|---|---|---|---|
| id | ||||||||
| 3 | 2023-06-15 | A | bias | prose | 600 | 600 | 1 | |
| 4 | 2023-06-15 | A | dark | prose | 600 | 600 | 1 | |
| 2 | 2023-06-15 | A | a | flat | prose | 600 | 600 | 4 |
| 1 | 2023-06-15 | A | a | light | prose | 600 | 600 | 100 |
We have 100 images of the prose target together with some calibration files.
Extracting photometry#
Using a reference#
In order to perform the photometric extraction of stars fluxes on all images, we will select a reference image from which we will extract:
The stars positions, then reused and detected on all images (after being aligned to the reference)
The global full-width at half-maximum (fwhm) of the PSF, to scale the apertures used in the aperture photometry block
from prose import FITSImage
ref = FITSImage(fm.all_images[0])
Note
FITSImage is the primary function used to load a FITS image into an Image object. While other loaders can be used, this loader is compatible with most of the images aquired from small to large-size ground-based telescopes.
Let’s now build a Sequence to process the reference image
from prose import Sequence, blocks
calibration = Sequence(
[
blocks.Calibration(darks=fm.all_darks, bias=fm.all_bias, flats=fm.all_flats),
blocks.Trim(),
blocks.PointSourceDetection(), # stars detection
blocks.Cutouts(21), # making stars cutouts
blocks.MedianEPSF(), # building PSF
blocks.psf.Moffat2D(), # modeling PSF
]
)
calibration.run(ref, show_progress=False)
ref.show()
ref.sources.plot()
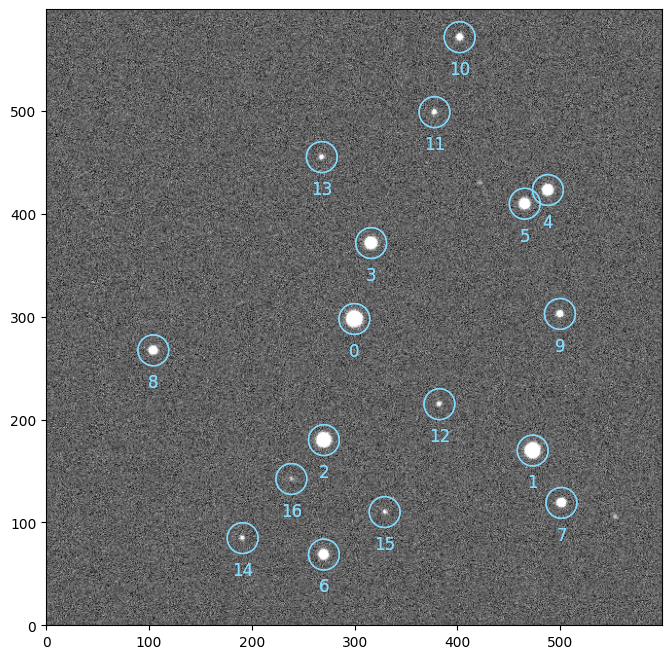
Tip
You can use a Gaia query to define which stars you want the photometry from (e.g. if too faint to be detected here)
Aperture photometry#
We can now extract the photometry of these stars
radii = np.linspace(0.2, 4, 30)
photometry = Sequence(
[
*calibration, # calibration
blocks.ComputeTransformTwirl(ref), # compute alignment
blocks.AlignReferenceSources(ref), # alignment
blocks.CentroidQuadratic(), # centroiding
blocks.AperturePhotometry(radii=radii), # aperture photometry
blocks.AnnulusBackground(), # local background estimate
blocks.GetFluxes(),
]
)
photometry.run(fm.all_images, loader=FITSImage)
Notice how we specified a loader to photometry.run, and instead of passing a list of Image objects, we passed a list of image paths. Although we cannot access each Image and its data individually, this allow to process a large amount of images without having to load them in memory. Instead, eadch image is loaded, processed and deleted in memory on the go.
The Fluxes object#
The GetFluxes block provide a way to retrieve the fluxes computed by the AperturePhotometry minus the background estimated with AnnulusBackground
raw_fluxes = photometry[-1].fluxes
we can then produce the differential photometry with autodiff
# picking target
raw_fluxes.target = 0
# good practice
raw_fluxes = raw_fluxes.sigma_clipping_data(bkg=3)
# differential photometry
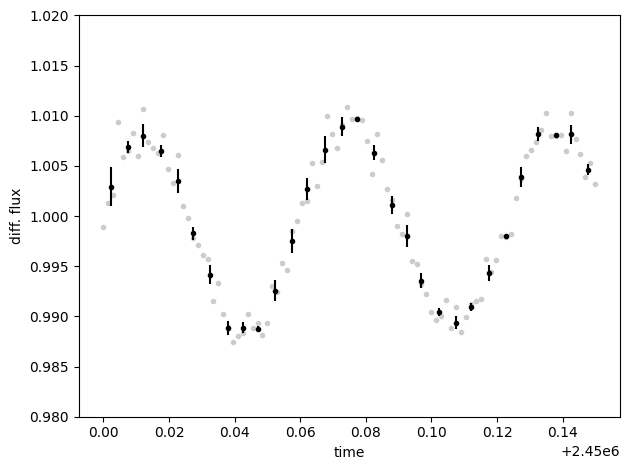
diff_fluxes = raw_fluxes.autodiff()
diff_fluxes.plot()
diff_fluxes.bin(0.005, True).errorbar()
plt.ylim(0.98, 1.02)
plt.xlabel("time")
plt.ylabel("diff. flux")
plt.tight_layout()
Show code cell content
import shutil
shutil.rmtree(fits_folder)